Machine learning for package users with R (4): Neural Network
These days almost everybody appears to love a variation of Neural Network (NN) -- Deep Learning. I already argued about how Deep Learning works and what kind of parameters characterizes it in the previous post.
A brief trial on a short version of MNIST datasets
In this post I argue about its origin, usual 3-layers (with only a hidden layer) NN.
> library(nnet) > train<-read.csv("short_prac_train.csv") > test<-read.csv("short_prac_test.csv") > train$label<-as.factor(train$label) > test$label<-as.factor(test$label) > train.nnet<-nnet(label~.,train,size=100,decay=0.01,maxit=100,MaxNWts=80000) # weights: 79510 initial value 21847.796288 iter 10 value 5304.553496 iter 20 value 3625.223363 iter 30 value 3286.108136 iter 40 value 2882.645985 iter 50 value 2544.534809 iter 60 value 2426.173862 iter 70 value 2300.468337 iter 80 value 2200.327282 iter 90 value 2100.783283 iter 100 value 2042.220475 final value 2042.220475 stopped after 100 iterations # took mmmmmmmmmmmmuch long time... > table(test$label,predict(train.nnet,newdata=test[,-1],type='class')) 0 1 2 3 4 5 6 7 8 9 0 93 0 1 0 0 2 2 0 2 0 1 0 93 1 0 0 2 0 2 2 0 2 3 3 83 3 1 0 1 2 4 0 3 2 0 3 79 0 7 1 2 4 2 4 0 1 1 0 86 2 3 0 1 6 5 2 0 0 6 2 79 4 1 4 2 6 2 0 3 0 4 3 87 0 0 1 7 1 1 1 0 1 0 0 88 2 6 8 0 0 4 2 1 8 0 2 81 2 9 0 0 1 2 6 0 0 4 4 83 > sum(diag(table(test$label,predict(train.nnet,newdata=test[,-1],type='class'))))/nrow(test) [1] 0.852 # OMG, it's low...
Not surprisingly, it's accuracy is low :P) I understand, in principle, NN is hard to be tuned and so that some people call it "a kind of master piece", that means only "masters" can tune it well.
Algorithm summary
As well known, NN is a perceptron with a hidden layer and "back propagation". Very fortunately, the famous textbook ESL clearly explains its algorithm at p.396 as below.
Here is back-propagation in detail for squared error loss. Let
, from (11.5) and let
. Then we have
(11.11)
with derivatives
(11.12)
Given these derivatives, a gradient descent update at the (r + 1)st iteration has the form
(11.13)
where
is the learning rate, discussed below.
Now write (11.12) as
(11.14)
The quantities
and
are "errors" from the current model at the output and hidden layer units, respectively. From their definitions, these errors satisfy
(11.15)
known as the back-propagation equations. Using this, the updates in (11.13) can be implemented with a two-pass algorithm. In the forward pass, the current weights are fixed and the predicted values
are computed from formula (11.5). In the backward pass, the errors
are computed, and then back-propagated via (11.15) to give the errors
. Both sets of errors are then used to compute the gradients for the updates in (11.13), via (11.14).
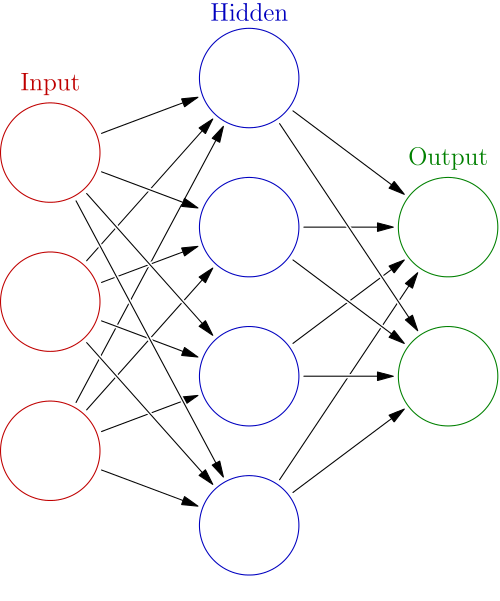
(wikipedia:en:Artificial_neural_networks)
This algorithm summary says that NN is rather an iterative optimizing procedure than "network". I guess many people regard NN as "network" but from the viewpoint of its algorithm it is merely a two-pass algorithm. This view will be important for understanding how Deep Learning works, as an extension of NN.
How it works on XOR patterns and linearly separable patterns
XOR patterns
OK, let's try it on XOR patterns. I think you already download simple and complex XOR patterns from my GitHub repository. First let's run a parameter tuning routine. Import {nnet} and {caret} packages and run as below.
> library(nnet) > library(caret) > xors<-read.table("xor_simple.txt",header=T) > xors$label<-as.factor(xors$label-1) > xorc<-read.table("xor_complex.txt",header=T) > xorc$label<-as.factor(xorc$label-1) # Tune for simple XOR pattern > xors.tune<-train(label~.,data=xors,method="nnet",tuneLength=4,maxit=100,trace=F) Loading required package: class > print(xors.tune) Neural Network 100 samples 2 predictors 2 classes: '0', '1' No pre-processing Resampling: Bootstrapped (25 reps) Summary of sample sizes: 100, 100, 100, 100, 100, 100, ... Resampling results across tuning parameters: size decay Accuracy Kappa Accuracy SD Kappa SD 1 0 0.659 0.326 0.06 0.099 1 1e-04 0.65 0.283 0.0466 0.0892 1 0.00316 0.645 0.284 0.0544 0.0934 1 0.1 0.643 0.296 0.0611 0.105 3 0 0.892 0.784 0.0773 0.149 3 1e-04 0.888 0.772 0.101 0.206 3 0.00316 0.924 0.846 0.0732 0.146 3 0.1 0.923 0.845 0.0694 0.139 5 0 0.913 0.825 0.0512 0.097 5 1e-04 0.933 0.865 0.0403 0.0802 5 0.00316 0.951 0.901 0.0405 0.0803 5 0.1 0.963 0.925 0.0304 0.0612 7 0 0.925 0.849 0.0394 0.0782 7 1e-04 0.951 0.901 0.0433 0.086 7 0.00316 0.95 0.9 0.0431 0.0846 7 0.1 0.961 0.921 0.0328 0.0657 Accuracy was used to select the optimal model using the largest value. The final values used for the model were size = 5 and decay = 0.1. # Tune for complex XOR pattern > xorc.tune<-train(label~.,data=xorc,method="nnet",tuneLength=4,maxit=100,trace=F) > print(xorc.tune) Neural Network 100 samples 2 predictors 2 classes: '0', '1' No pre-processing Resampling: Bootstrapped (25 reps) Summary of sample sizes: 100, 100, 100, 100, 100, 100, ... Resampling results across tuning parameters: size decay Accuracy Kappa Accuracy SD Kappa SD 1 0 0.521 0.0583 0.0808 0.153 1 1e-04 0.509 0.0332 0.0822 0.152 1 0.00316 0.531 0.0876 0.0839 0.158 1 0.1 0.519 0.0638 0.0879 0.176 3 0 0.652 0.302 0.0985 0.193 3 1e-04 0.667 0.333 0.0676 0.133 3 0.00316 0.678 0.359 0.0808 0.148 3 0.1 0.698 0.394 0.0609 0.119 5 0 0.677 0.349 0.08 0.161 5 1e-04 0.673 0.345 0.0758 0.145 5 0.00316 0.678 0.355 0.0628 0.126 5 0.1 0.715 0.43 0.079 0.152 7 0 0.685 0.371 0.0942 0.184 7 1e-04 0.68 0.361 0.067 0.135 7 0.00316 0.681 0.363 0.0672 0.131 7 0.1 0.715 0.431 0.0745 0.144 Accuracy was used to select the optimal model using the largest value. The final values used for the model were size = 7 and decay = 0.1.
Now we have a set of best parameters for both simple and complex XOR patterns.
# Set a grid > px<-seq(-4,4,0.03) > py<-seq(-4,4,0.03) > pgrid<-expand.grid(px,py) > names(pgrid)<-names(xors)[-3] # Simple XOR pattern > plot(c(),type='n',xlim=c(-4,4),ylim=c(-4,4)) > par(new=T) > rect(0,0,4,4,col='#aaaaff') > par(new=T) > rect(-4,0,0,4,col='#ffaaaa') > par(new=T) > rect(-4,-4,0,0,col='#aaaaff') > par(new=T) > rect(0,-4,4,0,col='#ffaaaa') > par(new=T) > plot(xors[,-3],col=c(rep('blue',50),rep('red',50)),xlim=c(-4,4),ylim=c(-4,4),pch=19,cex=2.5) > par(new=T) > contour(px,py,array(predict(xors.nnet,newdata=pgrid),dim = c(length(px),length(py))),levels = 0.5,drawlabels = T,col='purple',lwd=5,xlim=c(-4,4),ylim=c(-4,4)) # Complex XOR pattern > plot(c(),type='n',xlim=c(-4,4),ylim=c(-4,4)) > par(new=T) > rect(0,0,4,4,col='#aaaaff') > par(new=T) > rect(-4,0,0,4,col='#ffaaaa') > par(new=T) > rect(-4,-4,0,0,col='#aaaaff') > par(new=T) > rect(0,-4,4,0,col='#ffaaaa') > par(new=T) > plot(xorc[,-3],col=c(rep('blue',50),rep('red',50)),xlim=c(-4,4),ylim=c(-4,4),pch=19,cex=2.5) > par(new=T) > contour(px,py,array(predict(xorc.nnet,newdata=pgrid),dim = c(length(px),length(py))),levels = 0.5,drawlabels = T,col='purple',lwd=5,xlim=c(-4,4),ylim=c(-4,4))


Both look nice. Even in the case of complex XOR pattern, this NN model provides well-smoothed and generalized decision boundaries.
But we can arbitrarily excessively generalize or overfit it to complex XOR pattern with tuning in a bad manner. See below.
# Try to excessively generalize > xorc.nnet2<-nnet(label~.,xorc,size=2,decay=0.1) # weights: 9 initial value 69.974985 iter 10 value 68.637481 iter 20 value 61.866228 iter 30 value 58.071456 final value 58.067022 converged > plot(c(),type='n',xlim=c(-4,4),ylim=c(-4,4)) > par(new=T) > rect(0,0,4,4,col='#aaaaff') > par(new=T) > rect(-4,0,0,4,col='#ffaaaa') > par(new=T) > rect(-4,-4,0,0,col='#aaaaff') > par(new=T) > rect(0,-4,4,0,col='#ffaaaa') > par(new=T) > plot(xorc[,-3],col=c(rep('blue',50),rep('red',50)),xlim=c(-4,4),ylim=c(-4,4),pch=19,cex=2.5) > par(new=T) > contour(px,py,array(predict(xorc.nnet2,newdata=pgrid),dim = c(length(px),length(py))),levels = 0.5,drawlabels = T,col='purple',lwd=5,xlim=c(-4,4),ylim=c(-4,4)) # Try to overfit > xorc.nnet3<-nnet(label~.,xorc,size=30,decay=0.01,maxit = 500) # weights: 121 initial value 72.772471 iter 10 value 45.654101 iter 20 value 38.793337 iter 30 value 36.433649 iter 40 value 35.542237 iter 50 value 35.151096 iter 60 value 34.962654 iter 70 value 34.787609 iter 80 value 34.711248 iter 90 value 34.677120 iter 100 value 34.647901 iter 110 value 34.627443 iter 120 value 34.617480 iter 130 value 34.611521 iter 140 value 34.607867 iter 150 value 34.605730 iter 160 value 34.604129 iter 170 value 34.602038 iter 180 value 34.600587 iter 190 value 34.599829 iter 200 value 34.599381 iter 210 value 34.599158 iter 220 value 34.599070 iter 230 value 34.598967 iter 240 value 34.598846 iter 250 value 34.598784 iter 260 value 34.598687 iter 270 value 34.598562 iter 280 value 34.598544 final value 34.598544 converged > plot(c(),type='n',xlim=c(-4,4),ylim=c(-4,4)) > par(new=T) > rect(0,0,4,4,col='#aaaaff') > par(new=T) > rect(-4,0,0,4,col='#ffaaaa') > par(new=T) > rect(-4,-4,0,0,col='#aaaaff') > par(new=T) > rect(0,-4,4,0,col='#ffaaaa') > par(new=T) > plot(xorc[,-3],col=c(rep('blue',50),rep('red',50)),xlim=c(-4,4),ylim=c(-4,4),pch=19,cex=2.5) > par(new=T) > contour(px,py,array(predict(xorc.nnet3,newdata=pgrid),dim = c(length(px),length(py))),levels = 0.5,drawlabels = T,col='purple',lwd=5,xlim=c(-4,4),ylim=c(-4,4))

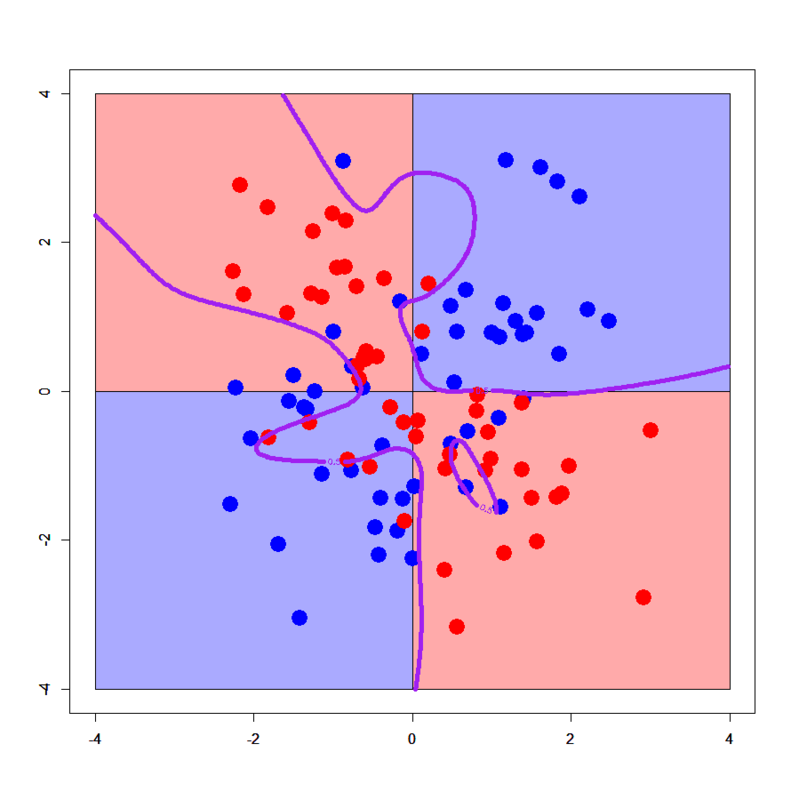
Hehe, looks crazy! :P) You can well find how NN can be easily tuned worse. So one of my previous colleagues who is an outstanding expert of NN told me "NN is a MASTER PIECE". Yes I agree it.
Linearly separable patterns
> dbi<-read.table("linear_bi.txt",header=T) > dbi$label<-as.factor(dbi$label) > summary(dbi) x y label Min. :0.2112 Min. :0.177 0:25 1st Qu.:1.0260 1st Qu.:1.133 1:25 Median :1.5830 Median :1.616 Mean :1.5786 Mean :1.606 3rd Qu.:2.0201 3rd Qu.:2.242 Max. :3.2749 Max. :2.994 > dml<-read.table("linear_multi.txt",header=T) > dml$label<-as.factor(dml$label) > px2<-seq(-0.5,4.5,0.03) > py2<-seq(-0.5,4.5,0.03) > pgrid2<-expand.grid(px2,py2) > names(pgrid2)<-names(dbi)[-3] > dbi.tune<-train(label~.,data=dbi,method="nnet",tuneLength=4,maxit=100,trace=F) > print(dbi.tune) Neural Network 50 samples 2 predictors 2 classes: '0', '1' No pre-processing Resampling: Bootstrapped (25 reps) Summary of sample sizes: 50, 50, 50, 50, 50, 50, ... Resampling results across tuning parameters: size decay Accuracy Kappa Accuracy SD Kappa SD 1 0 0.884 0.763 0.0766 0.165 1 1e-04 0.878 0.751 0.0798 0.171 1 0.00316 0.884 0.766 0.0695 0.145 1 0.1 0.878 0.756 0.0786 0.16 3 0 0.864 0.724 0.0644 0.129 3 1e-04 0.874 0.745 0.0722 0.144 3 0.00316 0.846 0.688 0.0792 0.16 3 0.1 0.881 0.762 0.0735 0.147 5 0 0.841 0.676 0.0874 0.179 5 1e-04 0.834 0.666 0.0984 0.198 5 0.00316 0.831 0.66 0.0797 0.159 5 0.1 0.874 0.746 0.0853 0.172 7 0 0.854 0.705 0.077 0.152 7 1e-04 0.829 0.653 0.0894 0.18 7 0.00316 0.818 0.634 0.0904 0.18 7 0.1 0.874 0.746 0.0853 0.172 Accuracy was used to select the optimal model using the largest value. The final values used for the model were size = 1 and decay = 0.00316. > dml.tune<-train(label~.,data=dml,method="nnet",tuneLength=4,maxit=100,trace=F) > print(dml.tune) Neural Network 75 samples 2 predictors 3 classes: '0', '1', '2' No pre-processing Resampling: Bootstrapped (25 reps) Summary of sample sizes: 75, 75, 75, 75, 75, 75, ... Resampling results across tuning parameters: size decay Accuracy Kappa Accuracy SD Kappa SD 1 0 0.81 0.711 0.0948 0.145 1 1e-04 0.828 0.74 0.0582 0.0855 1 0.00316 0.835 0.75 0.0568 0.0842 1 0.1 0.784 0.678 0.078 0.108 3 0 0.823 0.731 0.051 0.0763 3 1e-04 0.812 0.711 0.0601 0.0978 3 0.00316 0.82 0.728 0.0728 0.114 3 0.1 0.835 0.75 0.0544 0.0813 5 0 0.801 0.701 0.0701 0.103 5 1e-04 0.81 0.713 0.0601 0.0906 5 0.00316 0.802 0.7 0.0646 0.102 5 0.1 0.83 0.743 0.0494 0.0737 7 0 0.79 0.684 0.0752 0.113 7 1e-04 0.785 0.676 0.066 0.0974 7 0.00316 0.804 0.705 0.0629 0.0961 7 0.1 0.826 0.738 0.0575 0.0841 Accuracy was used to select the optimal model using the largest value. The final values used for the model were size = 3 and decay = 0.1. # Fit NNs > dbi.nnet<-nnet(label~.,dbi,size=1,decay=dbi.tune$bestTune$decay) # weights: 5 initial value 35.776034 iter 10 value 13.686940 iter 20 value 10.696310 iter 30 value 10.639279 iter 40 value 10.636601 final value 10.636598 converged > dml.nnet<-nnet(label~.,dml,size=3,decay=dml.tune$bestTune$decay) # weights: 21 initial value 96.421336 iter 10 value 47.021366 iter 20 value 38.167857 iter 30 value 37.854914 iter 40 value 37.735454 iter 50 value 37.730520 iter 60 value 37.729962 final value 37.729953 converged # 2-classes > plot(c(),type='n',xlim=c(-0.5,3.5),ylim=c(-0.5,3.5)) > par(new=T) > polygon(c(-0.5,-0.5,3.5),c(3.5,-0.5,-0.5),col='#aaaaff') > par(new=T) > polygon(c(-0.5,3.5,3.5),c(3.5,-0.5,3.5),col='#ffaaaa') > par(new=T) > plot(dbi[,-3],pch=19,col=c(rep('blue',25),rep('red',25)),cex=3,xlim=c(-0.5,3.5),ylim=c(-0.5,3.5)) > par(new=T) > contour(px2,py2,array(predict(dbi.nnet,newdata=pgrid2),dim = c(length(px2),length(py2))),xlim=c(-0.5,3.5),ylim=c(-0.5,3.5),lwd=6,col='purple',levels = 0.5,drawlabels = T) # 3-classes > plot(c(),type='n',xlim=c(-0.5,4.5),ylim=c(-0.5,4.5)) > par(new=T) > polygon(c(-0.5,-0.5,3.5),c(3.5,-0.5,-0.5),col='#aaaaff') > par(new=T) > polygon(c(-0.5,3.5,4.5,4.5,1.0,-0.5),c(3.5,-0.5,-0.5,1.0,4.5,4.5),col='#ffaaaa') > par(new=T) > polygon(c(1.0,4.5,4.5),c(4.5,1.0,4.5),col='#ccffcc') > par(new=T) > plot(dml[,-3],pch=19,col=c(rep('blue',25),rep('red',25),rep('green',25)),cex=3,xlim=c(-0.5,4.5),ylim=c(-0.5,4.5)) > par(new=T) > contour(px2,py2,array(predict(dml.nnet,newdata=pgrid2,type='class'),dim=c(length(px2),length(py2))),xlim=c(-0.5,4.5),ylim=c(-0.5,4.5),col="purple",lwd=6,drawlabels=T)

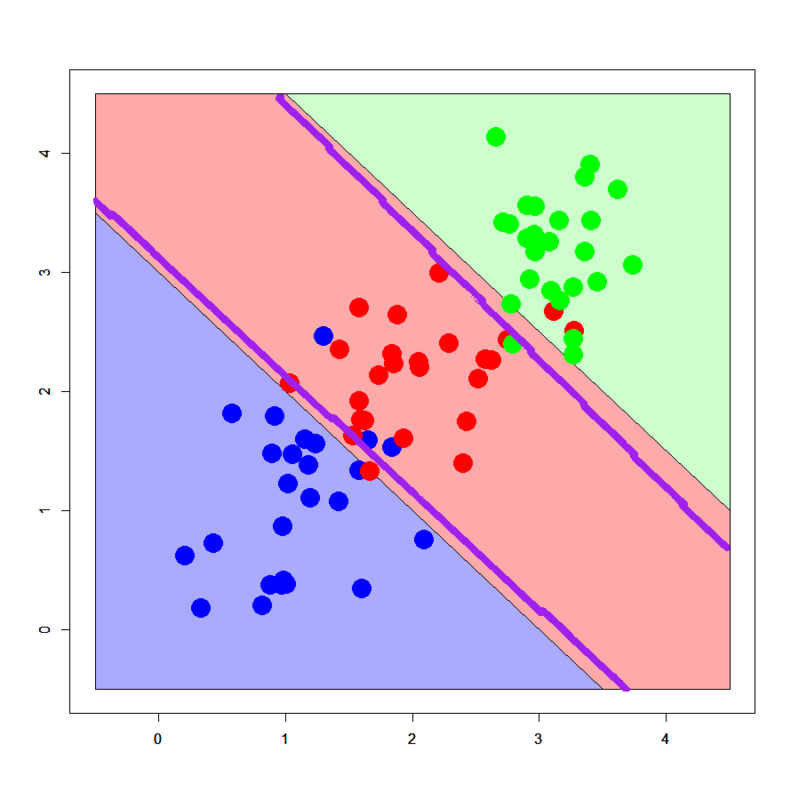
It's confirmed well-tuned NNs successfully draw decision boundaries along the assumed true ones. One of the advantages of NN is, I think, that it's extended from linear perceptron so it can also handle usual linearly separable patterns if correctly tuned.
Conclusions
Lessons we learned here are:
- NN with a single hidden layer is just an iterative optimized learning, rather than "network"
- Performance of NN can be easily affected by parameter tunings
- But NN also work well for both linearly separable and non-separable patterns
I feel these characteristics are taken over by Deep Learning and its family. In that sense, I think we have to seriously review NN with a single hidden layer.
Appendix
h2o.deeplearning() performed as below.
> library(h2o) > localH2O <- h2o.init(ip = "localhost", port = 54321, startH2O = TRUE, nthreads=7) > trData<-h2o.importFile(localH2O,path = "short_prac_train.csv") > tsData<-h2o.importFile(localH2O,path = "short_prac_test.csv") > res.dl <- h2o.deeplearning(x = 2:785, y = 1, data = trData, activation = "RectifierWithDropout",hidden=c(1024,1024,2048),epochs = 300, adaptive_rate = FALSE, rate=0.01, rate_annealing = 1.0e-6,rate_decay = 1.0, momentum_start = 0.5,momentum_ramp = 5000*18, momentum_stable = 0.99, input_dropout_ratio = 0.2,l1 = 1.0e-5,l2 = 0.0,max_w2 = 15.0, initial_weight_distribution = "Normal",initial_weight_scale = 0.01,nesterov_accelerated_gradient = T, loss = "CrossEntropy", fast_mode = T, diagnostics = T, ignore_const_cols = T,force_load_balance = T) > pred.dl<-h2o.predict(object=res.dl,newdata=tsData[,-1]) > pred.dl.df<-as.data.frame(pred.dl) > table(test$label,pred.dl.df[,1]) 0 1 2 3 4 5 6 7 8 9 0 95 0 1 1 0 0 3 0 0 0 1 0 99 1 0 0 0 0 0 0 0 2 0 0 97 0 1 0 0 1 0 1 3 0 0 1 96 0 2 0 1 0 0 4 0 1 1 0 96 0 2 0 0 0 5 0 2 0 3 0 94 1 0 0 0 6 1 0 0 0 0 1 98 0 0 0 7 0 0 0 0 3 0 0 97 0 0 8 0 0 1 3 1 2 0 1 92 0 9 0 0 0 0 4 0 0 2 0 94 > sum(diag(table(test$label,pred.dl.df[,1])))/nrow(test) [1] 0.958
Deep Learning worked a little better than random forest, as supposed to be shown in the next post. :D)